





















































































































The inherent safety and versatility of ultrasound imaging have made it widely accessible in modern clinical settings for disease diagnosis and health management. Artificial intelligence (AI) that can effectively learn ultrasound representations by integrating multi-source data holds significant promise for advancing clinical care. However, the scarcity of large labeled datasets in real-world clinical environments and the limited generalizability of task-specific models have hindered the development of generalizable clinical AI models for ultrasound applications. In this study, we present EchoCare, a novel ultrasound foundation model for generalist clinical use, developed via self-supervised learning on our curated, publicly available, large-scale unlabeled dataset EchoAtlas. EchoAtlas comprises 4.5 million ultrasound images, sourced from over 20 countries across 5 continents and acquired via a diverse range of distinct imaging devices, thus encompassing global cohorts that are multi-center, multi-device, and multi-ethnic. Unlike prior studies that adopt off-the-shelf vision foundation model architectures, we introduce a hierarchical classifier into EchoCare to enable joint learning of pixel-level and representation-level features, capturing both global anatomical contexts and local ultrasound characteristics. With minimal training, EchoCare outperforms state-of-the-art comparison models across 10 representative downstream ultrasound benchmarks of varying diagnostic difficulties, spanning disease diagnosis, lesion segmentation, organ detection, landmark prediction, quantitative regression, imaging enhancement and report generation. The code and pretrained model are publicly released, rendering EchoCare accessible for fine-tuning and local adaptation, supporting extensibility to additional applications. EchoCare provides a fully open and generalizable foundation model to boost the development of AI technologies for diverse clinical ultrasound applications.
EchoAtlas integrates multi-center, multi-region, and multi-device sources, covering 23 hospitals across 5 continents and 20 countries, ensuring diversity in clinical practices, patient demographics, and imaging equipment.
Hospitals Worldwide
Ultrasound Devices
Continents
Countries/Regions
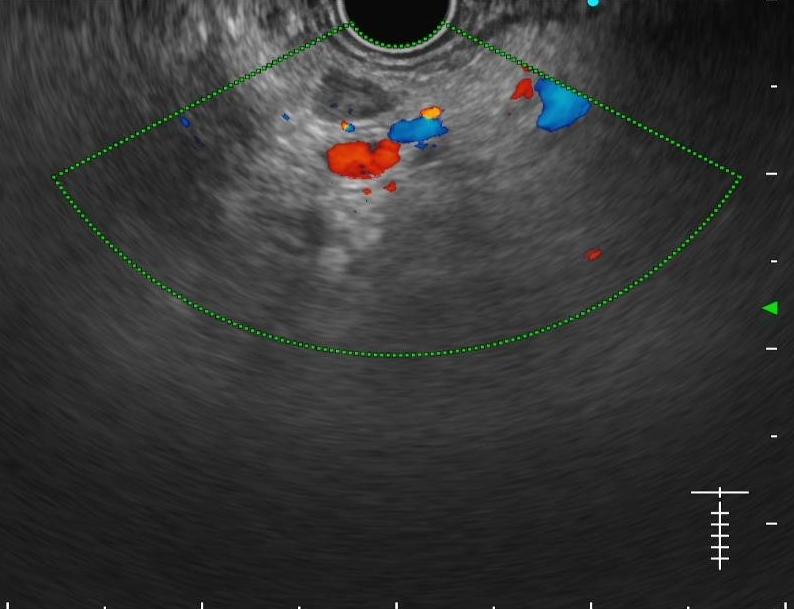
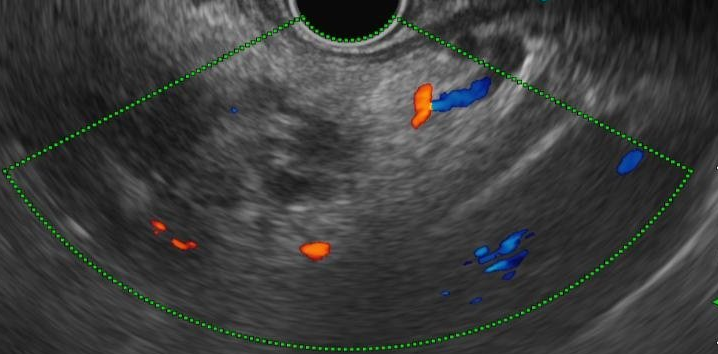
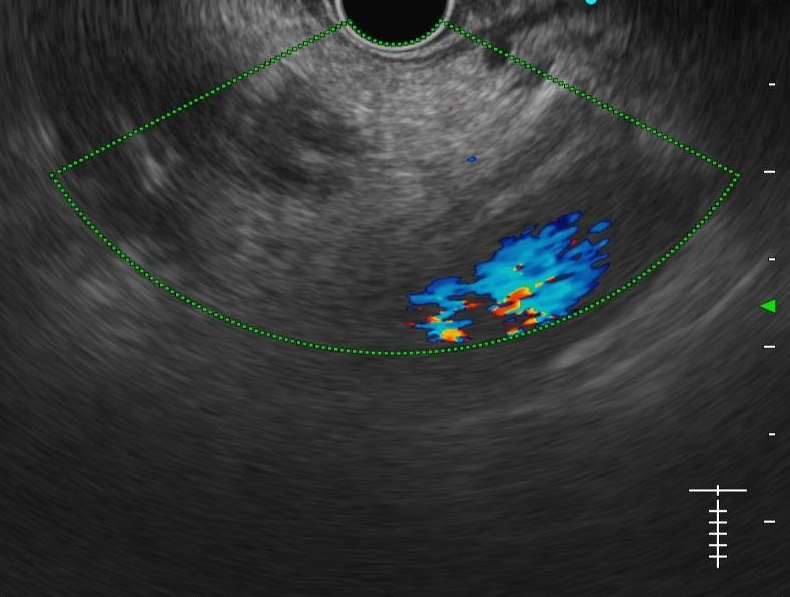
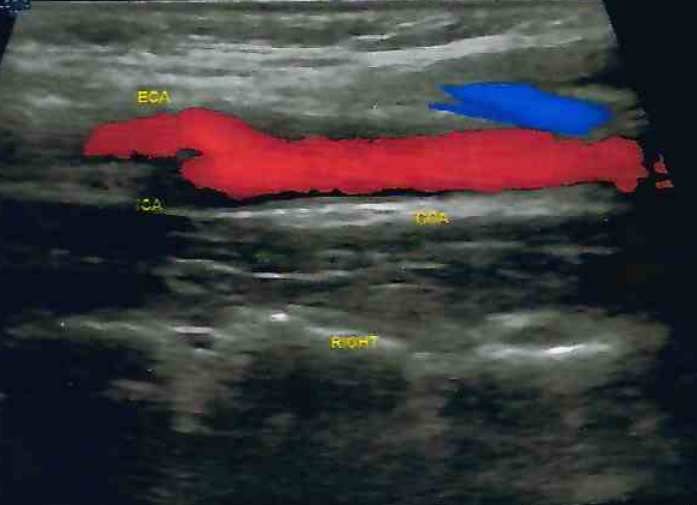
We evaluated different foundation models on three representative ultrasound clinical benchmarks for anatomical segmentation: the DDTI dataset for thyroid node segmentation, the Mus-V dataset for arterial-venous vessel segmentation, and the abdomen multi-organ segmentation.
Thyroid Node
Arterial-venous Vessel
Abdomen
Thyroid
Kidney
Liver
Breast
Carotid Artery
We evaluated EchoCare on the low-quality ultrasound image enhancement task using the USenhance benchmark dataset, which encompasses real-world clinical scans from 109 patients across five anatomical regions: thyroid, kidney, liver, breast, and carotid artery.
To evaluate the effectiveness of our developed foundation model in ultrasound report generation, we integrate EchoCare into an existing Transformer-based encoder–decoder report generator, where the input is the global visual features extracted from ultrasound images. The integrated model is then fine-tuned on the USData Liver dataset, which contains paired ultrasound images and corresponding expert-written reports.